Abstract
INTRODUCTION
C-X-C chemokine receptor type 7 (CXCR7) has recently been characterised as a novel receptor for the C-X-C motif chemokine 12 (CXCL12)/stromal cell-derived factor 1-alpha. CXCR7 has been thought to play an important role in the pathogenesis of chronic rhinosinusitis, angiogenesis and tumour metastasis. The present study aimed to examine the expression of CXCR7 in tissue samples of laryngeal cancer and maxillary sinus carcinoma to determine its role in the development of otorhinolaryngologic neoplasms.
METHODS
Samples of otorhinolaryngologic neoplasms were obtained from 17 patients with either nasal polyps (n = 7), laryngeal cancer (n = 5) or maxillary sinus carcinoma (n = 5), and who underwent surgical resection at West China Hospital of Sichuan University. Total RNA was isolated and CXCR7 mRNA expression was examined and quantified by relative real-time reverse transcription polymerase chain reaction. A one-way analysis of variance was performed using SPSS Statistics version 11.0 (SPSS Inc, Chicago, IL, USA) to compare the CXCR7 mRNA levels among the three groups of patients.
RESULTS
All samples tested positive for CXCR7 mRNA. The quantitative results showed that the CXCR7 mRNA levels were highest in laryngeal cancer and lowest in maxillary sinus carcinoma neoplasms, although there was no significant difference among the three samples.
CONCLUSION
CXCL12 and its receptor CXCR7 may contribute to eosinophilic inflammation in patients with chronic sinusitis and nasal polyps. Our results also suggest that CXCR7 may play a role in the progression, metastasis and angiogenesis of otorhinolaryngologic tumours.
INTRODUCTION
The C-X-C chemokine receptor type 7 (CXCR7) gene encodes a member of the G protein-coupled receptor family.(1) CXCR7, also known as an orphan receptor, was previously thought to be a receptor for vasoactive intestinal peptides.(2) It is now classified as a chemokine coreceptor, together with C-X-C chemokine receptor type 4 (CXCR4), for C-X-C motif chemokine 12/stromal cell-derived factor 1-alpha (CXCL12/SDF-1) and C-X-C motif chemokine I-TAC (CXCL11).(2) The receptors of CXCR12 are CXCR7 and CXCR4.
In vivo and in vitro investigations have demonstrated that CXCR7 is widely distributed in neoplasms such as lung and cervical cancer, and brain tumour tissues;(3-5) furthermore, a higher level of expression of CXCR7 is linked to an early recurrence rate as well as a lower five-year disease-free survival rate in lung cancer patients.(3) The molecular mechanism of CXCR7 signalling acts through the enhancement of environmental tolerance and proliferation of tumour cells. The binding of CXCL12 to CXCR7 has been reported to result in an increased in vitro tumour growth rate, whereas the introduction of a CXCR7 antagonist slows down the rate.(6)
Despite previous research on the tumourigenesis of several malignancies, the expression of CXCR7 in tissue samples of laryngeal cancer and maxillary sinus carcinoma has yet to be elucidated. The present study examined the expression of CXCR7 mRNA in tissue samples of laryngeal cancer and maxillary sinus carcinoma using real-time quantitative reverse transcriptase polymerase chain reaction (RT-PCR), which has not previously been reported; it was then compared to nasal polyp tissues in patients with chronic rhinosinusitis with nasal polyps (CRSwNPs) (benign lesions), which were taken as the positive control. We aimed to test the distinguished CXCR7 expression, in order to discuss the potential role of CXCR7 in otorhinolaryngologic neoplasms.
METHODS
This study was approved by the ethics committee of West China Hospital of Sichuan University, Chengdu, China. All participants provided verbal informed consent; written consent was not obtained as there was no existing standard for written consent in our centre. The otorhinolaryngologic neoplasm samples were removed via surgical resection. Demographic data (i.e. name, age, commodities, etc) from participants who provided verbal informed consent were recorded and included in our study.
Samples of otorhinolaryngologic neoplasms were obtained from 17 patients (ten women, seven men) with a mean age of 54.45 ± 11.25 years. Seven patients with nasal polyposis (two women, five men; mean age 43.43 ± 5.03 years) were also included. Five patients (two women, three men; mean age 65.22 ± 6.98 years) with laryngeal cancer, including one patient with well-differentiated squamous carcinoma, two patients with moderately differentiated squamous cell carcinoma and two patients with poorly differentiated squamous cell carcinoma, were chosen. Tissue samples of maxillary sinus carcinoma were taken from five patients (two women, three men; mean age 47.83 ± 9.54 years), including one patient with well-differentiated squamous carcinoma, two patients with moderately differentiated squamous cell carcinoma and two patients with poorly differentiated squamous cell carcinoma. All samples were from patients who underwent surgical resection of the tumours at West China Hospital of Sichuan University. All samples used for total RNA extraction were taken from within the tumours. Only tissues with anatomic pathology features that allowed for a matching diagnosis on magnetic resonance imaging and the pathology report were used.
In order to determine human CXCR7 mRNA levels, we designed primers for PCR amplification according to the NCBI GenBank (accession number NM_001047841). The primers were synthesised by Sangon Biotech Co Ltd®. The primers for human CXCR7 were as follows: forward primer 5’-CACTGCTACATCTTGAACCT-3’ and reverse primer 5’-GTTGATGGAGAAGATGAGGTGT-3’ [144 base pairs (bp)]. The primers for beta-actin (ACTB) were as follows: forward primer 5’-GAAGATCAAGATCATTGCTCCT-3’ and reverse primer 5’-TACTCCTGCTTGCTGATCCA-3’ (111 bp). TRIzol® reagent (Invitrogen, Grand Island, NY, USA) was used to isolate total mRNA from the tissues. A total of 5.0 µL of purified RNA was reverse transcribed into complementary DNA (cDNA) with the Revert Aid™ First Strand cDNA Synthesis Kit (MBI Fermentas Inc, Waltham, MA, USA) using random hexamer primers, as per the instructions. The cDNA was stored at –20°C until further analysis.
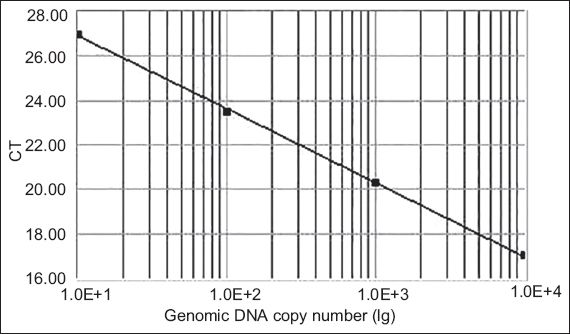
Real-time quantitative PCR was conducted to determine CXCR7 expression profiles in the collected tissues. The cDNAs were amplified using FTC-2000 real-time PCR machine (Funglyn Biotech, Toronto, Canada). A 30.0 µL reaction mixture consisting of 2.0 µL cDNA samples, 2.0 µL gene-specific forward and reverse primers (10.0 µM each), and 1.0 µL SYBR Green I dye was used for each analysis. Housekeeping gene ACTB was included to normalise the data. The amplification process was as follows: 45 cycles at 94ºC for two minutes, 94ºC for 20 seconds and 54ºC for 20 seconds, with a final extension at 60ºC for 30 seconds. The cycle threshold (Ct) number, which is defined as the number of PCR amplification cycles required for the sample to reach fluorescent intensity above the threshold, was determined for each gene and each developmental time point was analysed. Using serial dilutions of positive CXCR7 cDNA, a standard curve was generated on the basis of the linear relationship of existing Ct and the logarithm of the copy number. The slope of the curve was –3.30 and there was a strong linear relationship (R2.00;
Fig. 1
The standard curve for the CXCR7 set shows the cycle threshold (Ct) values against the genomic DNA copy number. Using serial dilutions of the standard cDNA, the standard curve ranged from 105 to 108 copies, generated on the basis of the linear relationship between the existing Ct and the logarithm of the copy number.
Results were normalised internally using the Ct number of the housekeeping gene ACTB as follows: ΔCt (sample) = (Ct sample) – (Ct ACTB). The mean Ct of CXCR7 mRNA from the blank negative control was set to a relative quantity (RQ) value of 1, using ΔΔCt that was calculated as follows: ΔΔCt (sample) = ΔCt (sample) – ΔCt (blank), and RQ = 2-ΔΔCt. The data was reported as mean ± standard deviation. A one-way analysis of variance was performed using SPSS version 11.0 (SPSS Inc, Chicago, IL, USA). A p-value < 0.05 was considered statistically significant.
RESULTS
CXCR7 mRNA was detected to be positive in all samples. The blank negative control, in which no cDNA was added, showed no positive reaction, indicating that there was no cross-contamination.
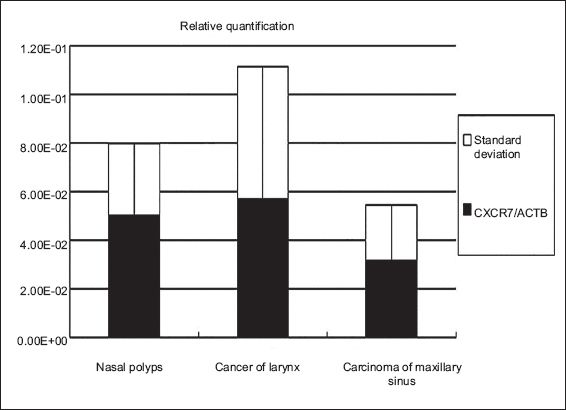
Expression of CXCR7 mRNA in tissue samples of nasal polyps, cancer of the larynx and carcinoma of the maxillary sinus was calculated according to the Ct value and normalised to the housekeeping gene, ACTB. The expression of CXCR7 mRNA was detected in all otorhinolaryngologic neoplasms examined in the current study; CXCR7 mRNA was slightly increased in tissues of cancer of the larynx and slightly decreased in carcinoma of the maxillary sinus as compared to the gene level of CXCR7 in the nasal polyps, which were the positive control. However, there was no statistical significance among the three groups (p > 0.05) (Figs.
Fig. 2
Northern blot shows ethidium bromide staining of polymerase chain reaction (PCR) CXCR7 cDNA in 2.0% agarose gel. Nasal polyp cDNA was used as a positive control for CXCR7 PCR primers (Lane 1). Amplification of CXCR7 cDNA produced a 144 base pair (bp) fragment, while amplification of beta-actin cDNA generated a 111 bp fragment. Lane 2: laryngeal cancer; Lane 3: maxillary sinus carcinoma; M: marker
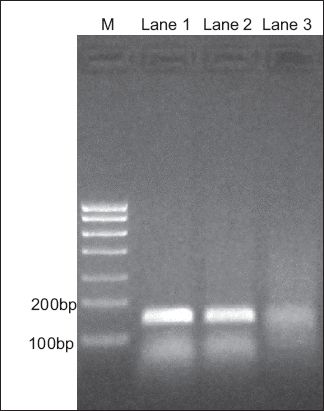
Fig. 3
The chart shows relative quantitative expression of CXCR7 mRNA in tissue samples from nasal polyps, laryngeal cancer and maxillary sinus carcinoma using real-time reverse transcriptase polymerase chain reaction.
DISCUSSION
In our study, CXCR7 was positive in otorhinolaryngologic tumours. The quantitative results showed that the levels of CXCR7 mRNA expression were highest in tissue samples from laryngeal cancer and lowest in those from maxillary sinus carcinoma, although there was no significant difference between these two groups; the results also showed that the expression levels of CXCR7 in otorhinolaryngologic malignant neoplasms of laryngeal cancer and maxillary sinus carcinoma were not significantly different from expression in nasal polyps, a benign neoplasm, which has not been reported before.
Our team previously reported that CXCR7 mRNA was widely distributed in many tumours, such as neurilemmoma, meningioma, pituitary adenoma and haemangioblastoma.(4) Our findings in the present study suggested that CXCR7 was also ubiquitously expressed in otorhinolaryngologic tumours, further supporting the idea that CXCR7 was associated with tumour proliferation, metastasis and angiogenesis in many other types of carcinoma.(7,8)
In maxillary sinus carcinoma tissue samples, the expression of CXCR7 was lowest in our study. A possible reason was that membrane-associated CXCR7 mRNA expression was specifically elevated in tumour-associated endothelial cells.(6) However, maxillary sinus carcinoma is a nonepithelial malignant neoplasm, composed of endothelial cells such as squamous epithelium, and glandular epithelial and nonepithelial cells (i.e. immune, neural and cartilage cells, and osteocytes). Therefore, the histology of maxillary sinus carcinoma was not purely endothelial cells that may have a higher level of expression of CXCR7 mRNA. As a high level of expression of CXCR7 indicates malignancy, CXCR7 may shed light on clinical features. Upregulation of CXCR7 expression could also aid in making an early diagnosis. In the future, treatment of primary nonepithelial malignant neoplasms through the targeting of CXCR7 could be explored.
Nasal polyps are polypoid masses arising mainly from the mucous membranes of the nose and paranasal sinus. The overgrowths of the mucosa frequently accompany eosinophilic inflammation (allergic rhinitis), in which CXCL12 interaction with its receptors, CXCR7 and CXCR4, might increase the migration or activation of B cells and their products and contribute to allergic rhinitis in patients with CRSwNP.(9,10,11) Nasal polyps arising from CRSwNP have a higher expression than chronic rhinosinusitis without nasal polyps or normal tissue, which was used as the positive control in the present study.(9)
In this study, laryngeal cancer neoplasms were speculated to have the highest expression of CXCR7 among the three neoplasms tested. Firstly, hypoxic preconditioning induced expression of CXCR7 mRNA in bone marrow-derived mesenchymal stem cells (MSCs).(12,13) CXCR7 also promoted the migration, adhesion and survival of MSCs, while inhibition of CXCR7 reduced the cell number of mouse neural progenitor cells.(12,13) The positive expression of CXCR7 in tissue samples of laryngeal cancer might also be due to hypoxia, which was the common environmental factor in patients. Secondarily, membrane-associated CXCR7 expression was elevated in tumour-associated endothelial cells.(9) Both mRNA and protein expression of CXCR7 was reported to be upregulated in human microvascular endothelial cells and endothelial hepatocellular carcinoma cells (HCCs) under hypoxic conditions.(14) This could influence cancer of the larynx, which is a malignant epithelial neoplasm. Thirdly, CXCR7 might also be implicated in tumourigenesis and angiogenesis of laryngeal carcinoma, as it was highly expressed in tumour-associated blood vessels in lung and breast cancer tissues, and the overexpression of CXCR7 has been shown to enhance lung metastases.(15) It was also shown that CXCR7 was selectively upregulated in hypoxic lungs in mice and the application of a CXCR7 antagonist reduced pulmonary vascular cell proliferation and remodelling.(16,17) Knockout of CXCR7 inhibited HCC invasion, adhesion, angiogenesis and reduction of tumour growth in the xenograft model.(18) Although the question of whether enhanced cell adhesion slows down cell migration is still under debate, our work supports the presumption that CXCR7 signals the stimulation of metastasis, invasion, and angiogenesis.
Several studies have suggested a positive relationship between CXCR7 and degree of malignancy. It was reported in metabolic flux analysis or quantitative analysis that the expression of CXCR7 was weak in normal tissues and strong in neoplasms of prostate cancer, pancreatic adenocarcinoma and other metastatic diseases.(1,19) In this study, it was shown that the levels of CXCR7 in otorhinolaryngologic neoplasms taken from patients with cancer of the larynx or carcinoma of the maxillary sinus were not significantly different from those with nasal polyps; this finding appears to contradict that of previous studies that correlated the expression level of CXCR7 with degree of malignancy.(1,19) This may be due to the fact that a higher expression of CXCR7 is often observed in tumour-associated endothelial cells,(6) and that maxillary sinus carcinoma is a nonepithelial malignant tumour, while nasal polyps and laryngocarcinoma are epithelial neoplasms. Therefore, different histological sources may have affected the expression of CXCR7, producing a positive result in all samples with no significant difference between them.
It was believed that CXCR7 acts through the mitogen-activated protein kinase pathway to increase cell survival, but much remains unknown about the CXCR7 mechanism.(20) In conclusion, further study needs to be carried out on the mechanism through which CXCR7 acts and to determine the possibilities of CXCR7 as an early marker for tumour diagnosis.
ACKNOWLEDGEMENTS
The authors would like to thank Mr Zengliang Xia and Mr Faqiang Zhang of the West China Laboratory of Molecular Genetics, Sichuan University, Chengdu, China, for their invaluable technical assistance.


